Establishing Wikidata as the central hub for linked open life science data
Jens Ohlig
22. October 2014
Thanks to the amazing work of the Wikidata community, every human gene (according to the United States National Center for Biotechnology Information) now has a representative entity on Wikidata. We hope that these are the seeds for some amazing applications in biology and medicine. Here is a report from Benjamin Good, Andrew Su, and Andra Waagmeester on their work with Wikidata. Their work was supported by the National Institutes of Health under grant GM089820.
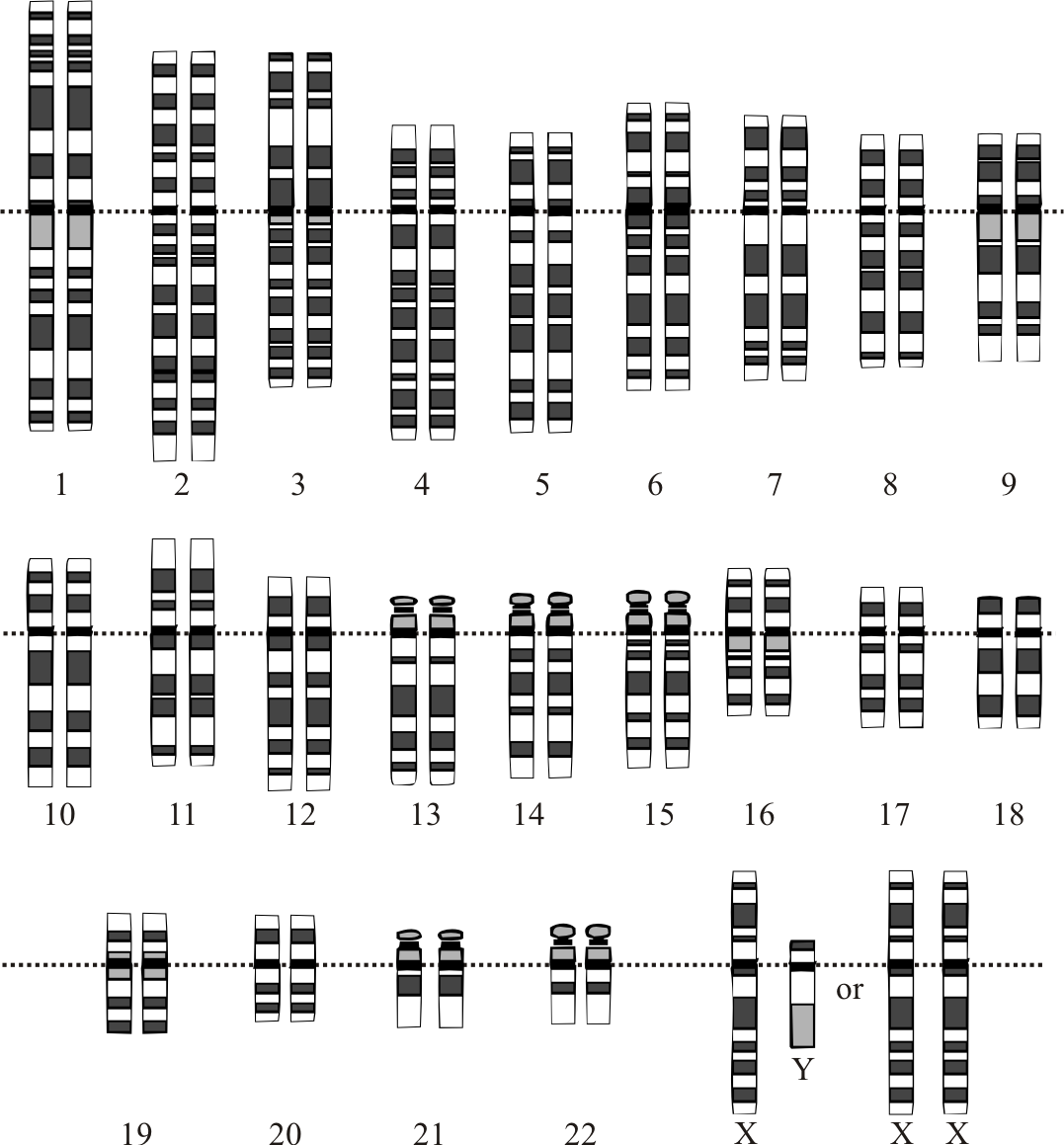
Graphical representation of the idealized human diploid karyotype, showing the organization of the genome into chromosomes. This drawing shows both the female (XX) and male (XY) versions of the 23rd chromosome pair. By Courtesy: National Human Genome Research Institute [Public domain], via Wikimedia Commons
In parallel to the appearance of Big Data in biology (and elsewhere), Wikipedia has arisen as one of the most important sources of all information on the Web. Within the context of Wikipedia, members of our research team have helped to foster the growth of a large collection of articles that describe the function and importance of human genes. Wikipedia and the subset of it that focuses on human genes (which we call the Gene Wiki), have flourished due to their centrality, the presence of the edit button, and the desire of the larger community to share knowledge openly.
Now, we are working to see if Wikidata can be the bridge between the open community-driven power of Wikipedia and the structured world of semantic data integration. Can the presence of that edit button on a centralized knowledge base associated with Wikipedia help the semantic web break through into everyday use within our community? The steps we are planning to take to test this idea within the context of the life sciences, are:
- Establishing bots that populate Wikidata with entities representative of three key classes: genes, diseases, and drugs.
- Expanding the scope of these bots to include the addition of statements that link these entities together into a valuable network of knowledge.
- Developing applications that display this information to the public that both encourage and enable them to contribute their knowledge back to Wikidata. The first implementation will be to use the Wikidata information to enhance the articles in Wikipedia.
We are excited to announce that the first step on this path has been completed!
Milestone 1 complete: Wikidata entities for all human genes
We have just finished adding entities for all (~40,000) human gene entries that contain a label in the Entrez Gene database [3]. We consider this a first step in making Wikidata a central repository for biological identifiers and their mappings to related resources. Each gene now has a stable identifier (e.g. https://www.wikidata.org/wiki/Q18039760) from Wikidata that links to other key identifier systems such as Entrez Gene. These are the first seeds of the garden of knowledge that we hope to sow.
We added Entrez Gene to Wikidata in two development sprints (see here and here). The final bot was split into two subtasks. The first being a stub creator, where for each Entrez Gene entry, a stub was created. This stub consisted of the title of the gene, its aliases, its Entrez Gene identifier (P351) it being a subclass of a gene (P279) and that it was found in taxon human (P703). After all the genes had their respective stubs, the second bot was run that enriched each entry with related identifiers and chromosomal positions (e.g. Ensembl Gene ID (P594), Ensembl Transcript ID (P704), HGNC ID (P354)). Although our first effort took approximately one month to complete all edits, we are currently optimizing this workflow based on valuable feedback from the Wikidata community. In the next days we will also use this approach to add Wikidata items for all mouse genes.
The development of the bot is organized around monthly sprints. In the current sprint, together with optimizing the current bot, diseases will be added. This will allow adding gene-disease links in Wikidata. We gladly welcome feedback, ideas and contributions to our bots!
The future
We look forward to an exciting time working together with the Wikidata community to change the way that biomedical data is managed towards a more efficient, more open model that results in more rapid advances in our overall understanding of biology and medicine. Join us on the mailing list, on WikiProject Molecular Biology, WikiProject Medicine and our open source code repository for Wikidata bots to help figure out how to make this happen!





[…] "Thanks to the amazing work of the Wikidata community, every human gene (according to the United States National Center for Biotechnology Information) now has a representative entity on Wikidata. We hope that these are the seeds for some amazing applications in biology and medicine. Here is a report from Benjamin Good, Andrew Su, and Andra Waagmeester on their work with Wikidata. Their work was supported by the National Institutes of Health under grant GM089820". […]
[…] […]